Ring filter#
Calculations#
For a ring resonator we need to define:
Optical parameters:
coupling coefficient: will define resonance extinction ratio for a particular ring loss.
Free spectral range.
Electrical parameters:
VpiL
Resistance
import gdsfactory as gf
import numpy as np
def ring(
wl: np.ndarray,
wl0: float,
neff: float,
ng: float,
ring_length: float,
coupling: float,
loss: float,
) -> np.ndarray:
"""Returns Frequency Domain Response of an all pass filter.
Args:
wl: wavelength in um.
wl0: center wavelength at which neff and ng are defined.
neff: effective index.
ng: group index.
ring_length: in um.
coupling: coupling coefficient.
loss: dB/um.
"""
transmission = 1 - coupling
neff_wl = (
neff + (wl0 - wl) * (ng - neff) / wl0
) # we expect a linear behavior with respect to wavelength
out = np.sqrt(transmission) - 10 ** (-loss * ring_length / 20.0) * np.exp(
2j * np.pi * neff_wl * ring_length / wl
)
out /= 1 - np.sqrt(transmission) * 10 ** (-loss * ring_length / 20.0) * np.exp(
2j * np.pi * neff_wl * ring_length / wl
)
return abs(out) ** 2
if __name__ == "__main__":
import matplotlib.pyplot as plt
loss = 0.03 # [dB/μm] (alpha) waveguide loss
neff = 2.46 # Effective index of the waveguides
wl0 = 1.55 # [μm] the wavelength at which neff and ng are defined
radius = 5
ring_length = 2 * np.pi * radius # [μm] Length of the ring
coupling = 0.5 # [] coupling of the coupler
wl = np.linspace(1.5, 1.6, 1000) # [μm] Wavelengths to sweep over
wl = np.linspace(1.55, 1.60, 1000) # [μm] Wavelengths to sweep over
ngs = [4.182551, 4.169563, 4.172917]
thicknesses = [210, 220, 230]
# widths = np.array([0.4, 0.45, 0.5, 0.55, 0.6])
# ngs = np.array([4.38215238, 4.27254985, 4.16956338, 4.13283219, 4.05791982])
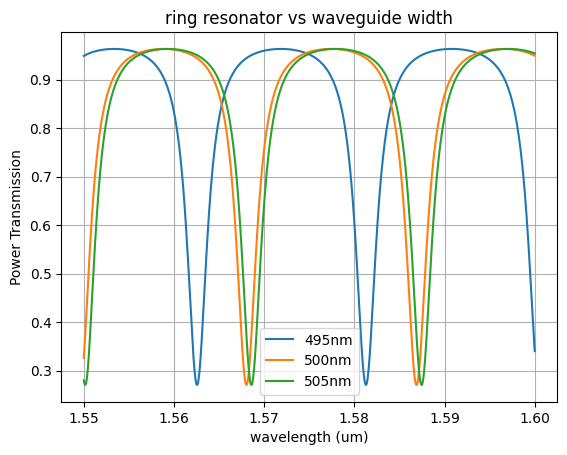
widths = np.array([0.495, 0.5, 0.505])
neffs = np.array([2.40197253, 2.46586378, 2.46731758])
ng = 4.2 # Group index of the waveguides
for width, neff in zip(widths, neffs):
p = ring(
wl=wl,
wl0=wl0,
neff=neff,
ng=ng,
ring_length=ring_length,
coupling=coupling,
loss=loss,
)
plt.plot(wl, p, label=f"{int(width*1e3)}nm")
plt.title("ring resonator vs waveguide width")
plt.xlabel("wavelength (um)")
plt.ylabel("Power Transmission")
plt.grid()
plt.legend()
plt.show()
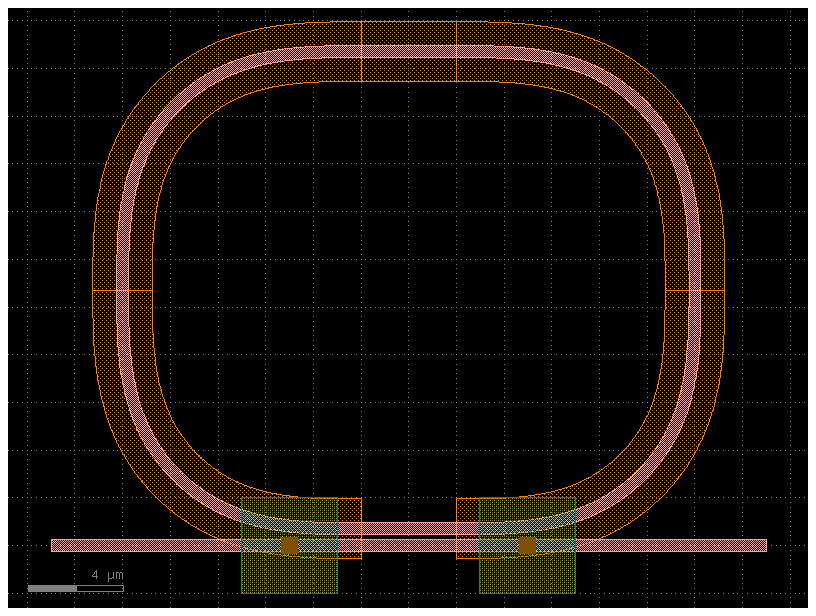
Layout#
gdsfactory easily enables you to layout Component with as many levels of hierarchy as you need.
A Component is a canvas where we can add polygons, references to other components or ports.
Lets add two references in a component.
c = gf.components.ring_single_heater(gap=0.2, radius=10, length_x=4)
c.show()
c.plot()
scene = c.to_3d()
scene.show()
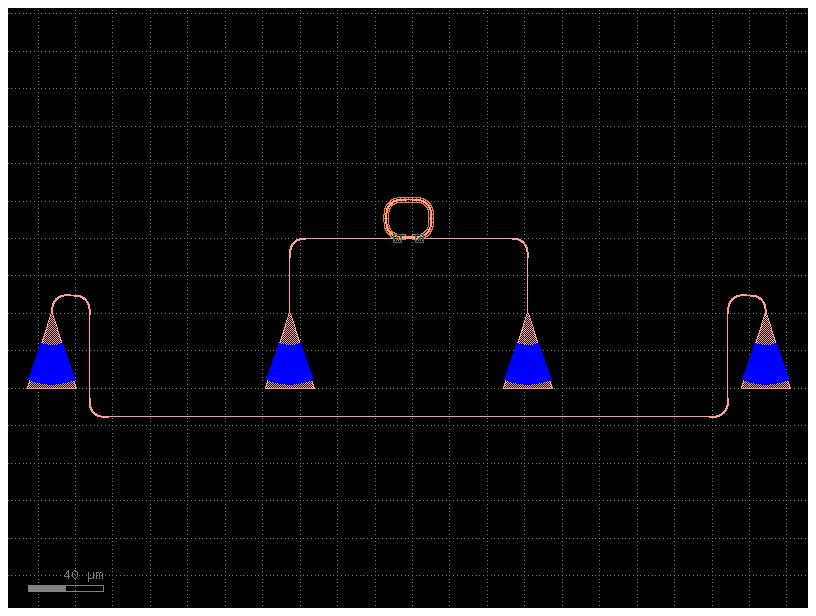
Lets define a ring function that also accepts other component specs for the subcomponents (straight, coupler, bend)
ring = gf.components.ring_single_heater(
gap=0.2, radius=10, length_x=4, via_stack_offset=(2, 0)
)
ring_with_grating_couplers = gf.routing.add_fiber_array(ring)
ring_with_grating_couplers.plot()
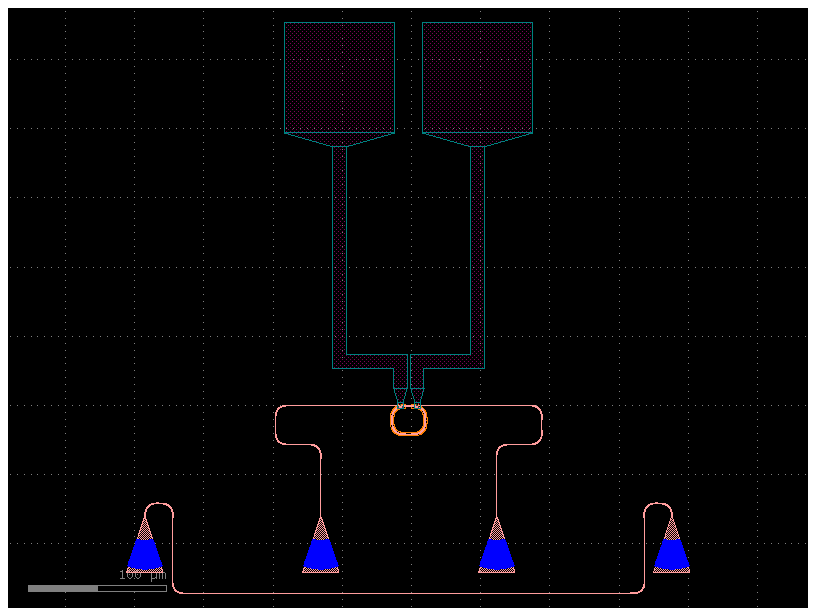
port_names = ["l_e1", "r_e3"]
port_names = ["l_e4", "r_e4"]
c = gf.routing.add_pads_top(ring, port_names=port_names, fanout_length=200)
c = gf.routing.add_fiber_array(c)
c
Top reticle assembly#
Once you have your components and circuits defined, you can add them into a top reticle Component for fabrication.
You need to consider:
what design variations do you want to include in the mask? You need to define your Design Of Experiment or DOE
obey DRC (Design rule checking) foundry rules for manufacturability. Foundry usually provides those rules for each layer (min width, min space, min density, max density)
make sure you will be able to test te devices after fabrication. Obey DFT (design for testing) rules. For example, if your test setup works only for fiber array, what is the fiber array spacing (127 or 250um?)
if you plan to package your device, make sure you follow your packaging guidelines from your packaging house (min pad size, min pad pitch, max number of rows for wire bonding …)
import gdsfactory as gf
import pandas as pd
@gf.cell
def spiral_gc(**kwargs):
"""Returns spiral with Grating Couplers."""
c = gf.components.spiral(**kwargs)
c = gf.routing.add_fiber_array(c)
c.info["doe"] = "spirals_sc" # strip Cband spirals
c.info["measurement"] = "optical_spectrum"
c.info["measurement_parameters"] = ""
c.info["analysis"] = "[power_envelope]"
c.info["analysis_parameters"] = "[]"
c.info["ports_optical"] = 4
c.info["ports_electrical"] = 0
c.info.update(kwargs)
return c
@gf.cell
def mzm_gc(length_x=10, **kwargs) -> gf.Component:
"""Returns a MZI with Grating Couplers.
Args:
length_x: length of the MZI.
kwargs: additional settings.
"""
c = gf.components.mzi2x2_2x2_phase_shifter(
length_x=length_x, auto_rename_ports=False, **kwargs
)
c = gf.routing.add_pads_top(c, port_names=["top_l_e1", "top_r_e3"])
c = gf.routing.add_fiber_array(c)
c.info["doe"] = "mzm"
c.info["measurement"] = "optical_spectrum"
c.info["analysis"] = "[fsr]"
c.info["analysis_parameters"] = "[]"
c.info["measurement_parameters"] = ""
c.info["ports_electrical"] = 2
c.info["ports_optical"] = 6
c.info["length_x"] = length_x
c.info.update(kwargs)
return c
def sample_reticle(grid: bool = False) -> gf.Component:
"""Returns MZI with TE grating couplers."""
mzis = [mzm_gc(length_x=lengths) for lengths in [100, 200, 300]]
spirals = [spiral_gc(length=length) for length in [0, 100, 200]]
rings = []
for length_x in [10, 20, 30]:
ring = gf.components.ring_single_heater(length_x=length_x)
c = gf.c.add_fiber_array_optical_south_electrical_north(
component=ring,
electrical_port_names=["l_e2", "r_e2"],
pad=gf.c.pad,
grating_coupler=gf.c.grating_coupler_te,
cross_section_metal="metal3",
)
c.name = f"ring_{length_x}"
c.info["doe"] = "ring_length_x"
c.info["measurement"] = "optical_spectrum"
c.info["measurement_parameters"] = ""
c.info["ports_electrical"] = 2
c.info["ports_optical"] = 4
c.info["analysis"] = "[fsr]"
c.info["analysis_parameters"] = "[]"
c.info["length_x"] = length_x
rings.append(c)
copies = 3 # number of copies of each component
components = mzis * copies + rings * copies + spirals * copies
if grid:
return gf.grid(components)
c = gf.pack(components)
if len(c) > 1:
c = gf.pack(c)[0]
return c[0]
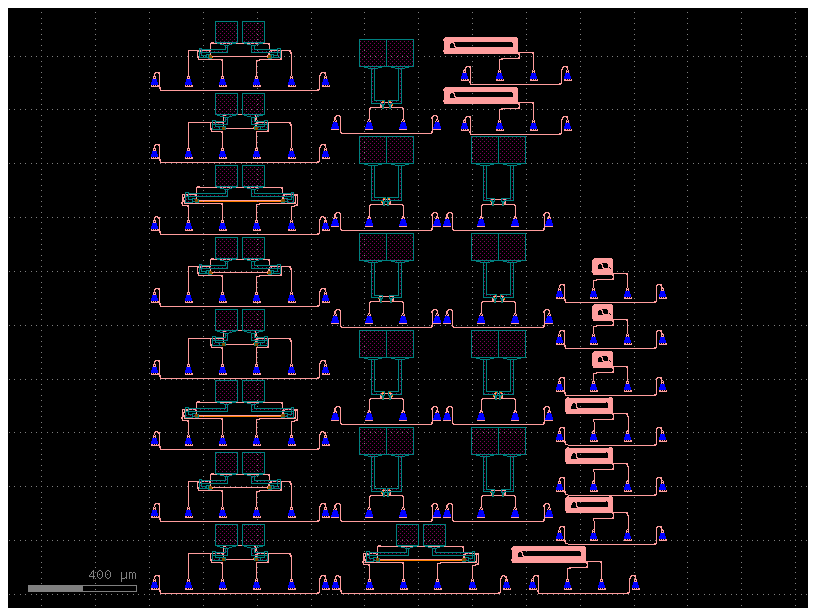
c = sample_reticle()
c.show()
c
gf.labels.write_test_manifest(c, csvpath="sample_reticle.csv")
df = pd.read_csv("sample_reticle.csv")
df
| cell | x | y | info | ports | settings | doe | analysis | analysis_parameters | measurement | measurement_parameters | ports_optical | ports_electrical | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | mzm_gc_LX100 | 250.020 | 153.225 | {"length_x": 100} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":100} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 1 | mzm_gc_LX200 | 200.020 | 419.775 | {"length_x": 200} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":200} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 2 | mzm_gc_LX300 | 150.020 | 686.325 | {"length_x": 300} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":300} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 3 | mzm_gc_LX100 | 250.020 | 952.876 | {"length_x": 100} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":100} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 4 | mzm_gc_LX200 | 200.020 | 1219.426 | {"length_x": 200} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":200} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 5 | mzm_gc_LX300 | 150.020 | 1485.976 | {"length_x": 300} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":300} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 6 | mzm_gc_LX100 | 250.020 | 1752.526 | {"length_x": 100} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":100} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 7 | mzm_gc_LX200 | 200.020 | 2019.076 | {"length_x": 200} | {"in_o1": {"name": "in_o1", "center": [145.02,... | {"length_x":200} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 8 | mzm_gc_LX300 | 820.960 | 153.225 | {"length_x": 300} | {"in_o1": {"name": "in_o1", "center": [815.96,... | {"length_x":300} | mzm | [fsr] | [] | optical_spectrum | NaN | 6 | 2 |
| 9 | ring_10 | 884.460 | 367.151 | {"length_x": 10} | {"o1": {"name": "o1", "center": [815.96, 300.6... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 10 | ring_20 | 889.460 | 727.151 | {"length_x": 20} | {"o1": {"name": "o1", "center": [815.96, 660.6... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 11 | ring_30 | 894.460 | 1087.151 | {"length_x": 30} | {"o1": {"name": "o1", "center": [815.96, 1020.... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 12 | ring_10 | 884.460 | 1447.151 | {"length_x": 10} | {"o1": {"name": "o1", "center": [815.96, 1380.... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 13 | ring_20 | 889.460 | 1807.151 | {"length_x": 20} | {"o1": {"name": "o1", "center": [815.96, 1740.... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 14 | ring_30 | 1311.400 | 367.151 | {"length_x": 30} | {"o1": {"name": "o1", "center": [1232.9, 300.6... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 15 | ring_10 | 1301.400 | 727.151 | {"length_x": 10} | {"o1": {"name": "o1", "center": [1232.9, 660.6... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 16 | ring_20 | 1306.400 | 1087.151 | {"length_x": 20} | {"o1": {"name": "o1", "center": [1232.9, 1020.... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 17 | ring_30 | 1311.400 | 1447.151 | {"length_x": 30} | {"o1": {"name": "o1", "center": [1232.9, 1380.... | {} | ring_length_x | [fsr] | [] | optical_spectrum | NaN | 4 | 2 |
| 18 | spiral_gc_L200 | 1395.179 | 140.351 | {"length": 200} | {"o1": {"name": "o1", "center": [1551.179, 34.... | {"length":200} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 19 | spiral_gc_L200 | 1141.179 | 1846.901 | {"length": 200} | {"o1": {"name": "o1", "center": [1297.179, 174... | {"length":200} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 20 | spiral_gc_L200 | 1141.179 | 2030.451 | {"length": 200} | {"o1": {"name": "o1", "center": [1297.179, 192... | {"length":200} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 21 | spiral_gc_L100 | 1593.840 | 323.901 | {"length": 100} | {"o1": {"name": "o1", "center": [1649.84000000... | {"length":100} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 22 | spiral_gc_L100 | 1593.840 | 507.451 | {"length": 100} | {"o1": {"name": "o1", "center": [1649.84000000... | {"length":100} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 23 | spiral_gc_L100 | 1593.840 | 691.001 | {"length": 100} | {"o1": {"name": "o1", "center": [1649.84000000... | {"length":100} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 24 | spiral_gc_L0 | 1693.840 | 864.301 | {"length": 0} | {"o1": {"name": "o1", "center": [1649.84000000... | {"length":0} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 25 | spiral_gc_L0 | 1693.840 | 1037.601 | {"length": 0} | {"o1": {"name": "o1", "center": [1649.84000000... | {"length":0} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |
| 26 | spiral_gc_L0 | 1693.840 | 1210.901 | {"length": 0} | {"o1": {"name": "o1", "center": [1649.84000000... | {"length":0} | spirals_sc | [power_envelope] | [] | optical_spectrum | NaN | 4 | 0 |