Components with hierarchy#
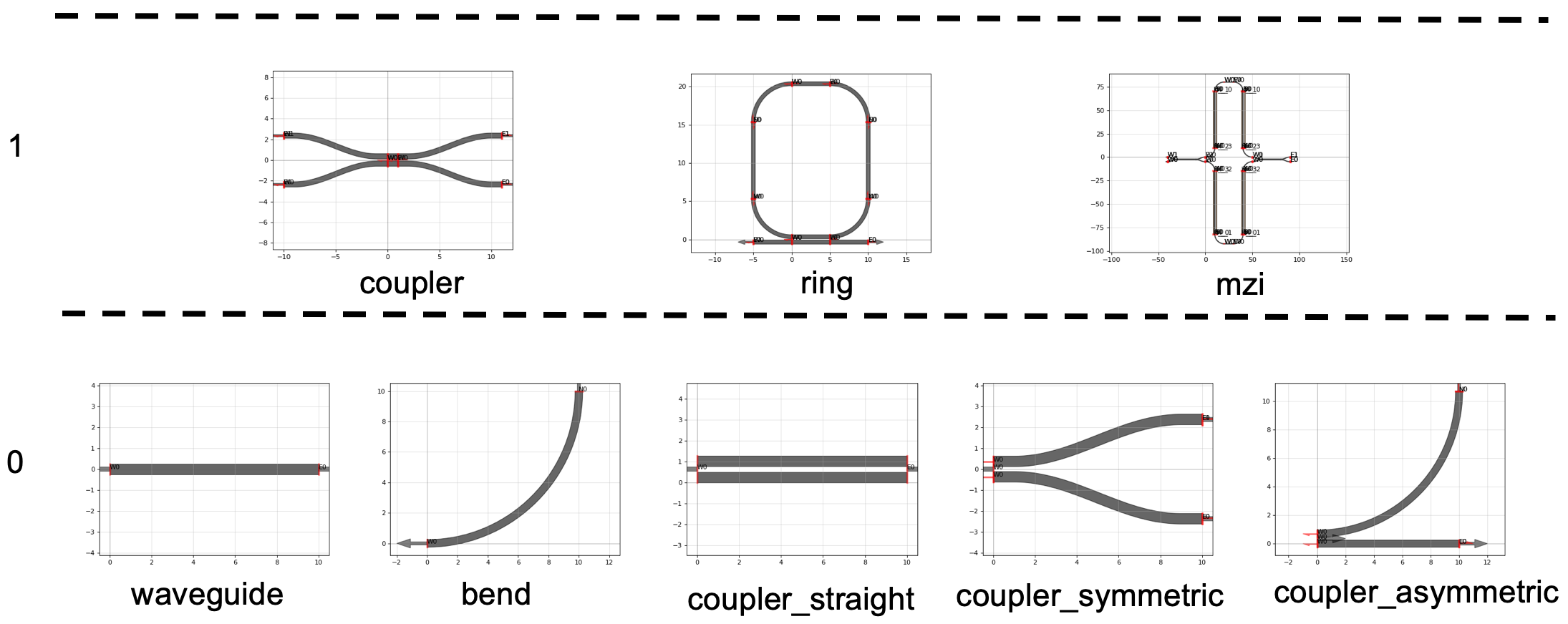
You can define component parametric cells (waveguides, bends, couplers) as functions with basic input parameters (width, length, radius …) and use them as arguments for composing more complex functions.
from functools import partial
import toolz
import gdsfactory as gf
from gdsfactory.typings import ComponentSpec
from gdsfactory.cross_section import CrossSectionSpec
gf.gpdk.PDK.activate()
Problem
When using hierarchical cells where you pass N subcells with M parameters you can end up with N*M parameters. This will make it hard to read the code.
@gf.cell
def bend_with_straight_with_too_many_input_parameters(
bend=gf.components.bend_euler,
straight=gf.components.straight,
length: float = 3,
angle: float = 90.0,
p: float = 0.5,
with_arc_floorplan: bool = True,
npoints: int | None = None,
cross_section: CrossSectionSpec = "strip",
) -> gf.Component:
"""As hierarchical cells become more complex, the number of input parameters can increase significantly."""
c = gf.Component()
b = bend(
angle=angle,
p=p,
with_arc_floorplan=with_arc_floorplan,
npoints=npoints,
cross_section=cross_section,
)
s = straight(length=length, cross_section=cross_section)
bref = c << b
sref = c << s
sref.connect("o2", bref.ports["o2"])
c.info["length"] = b.info["length"] + s.info["length"]
return c
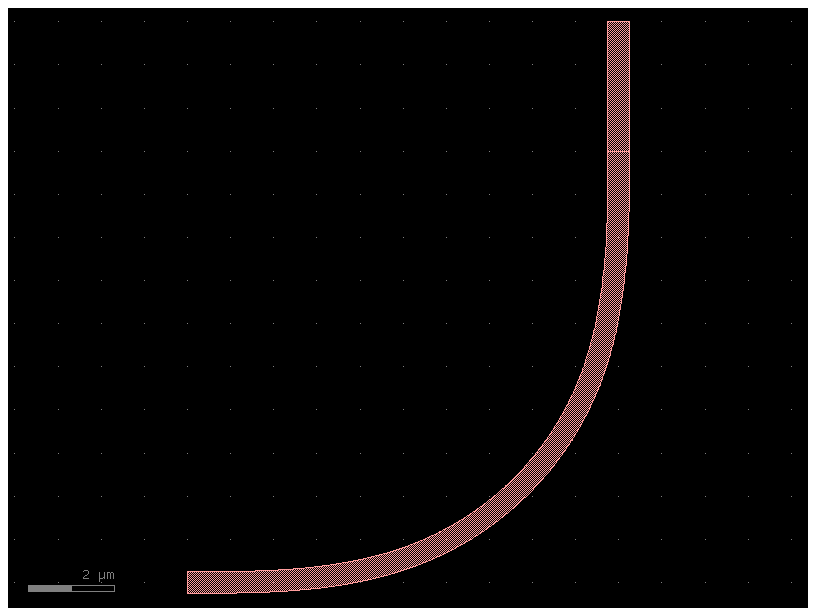
c = bend_with_straight_with_too_many_input_parameters()
c.plot()
Solution
You can use a ComponentSpec parameter for every subcell. The ComponentSpec can be a dictionary with arbitrary number of settings, a string, or a function.
ComponentSpec#
When defining a Parametric cell you can use other ComponentSpec as an argument. It can be a:
string: function name of a cell registered on the active PDK.
"bend_circular".dict:
dict(component='bend_circular', settings=dict(radius=20)).function: Using
functools.partialyou can customize the default parameters of a function.
@gf.cell
def bend_with_straight(
bend: ComponentSpec = gf.components.bend_euler,
straight: ComponentSpec = gf.components.straight,
) -> gf.Component:
"""Much simpler version.
Args:
bend: input bend.
straight: output straight.
"""
c = gf.Component()
b = gf.get_component(bend)
s = gf.get_component(straight)
bref = c << b
sref = c << s
sref.connect("o2", bref.ports["o2"])
c.info["length"] = b.info["length"] + s.info["length"]
return c
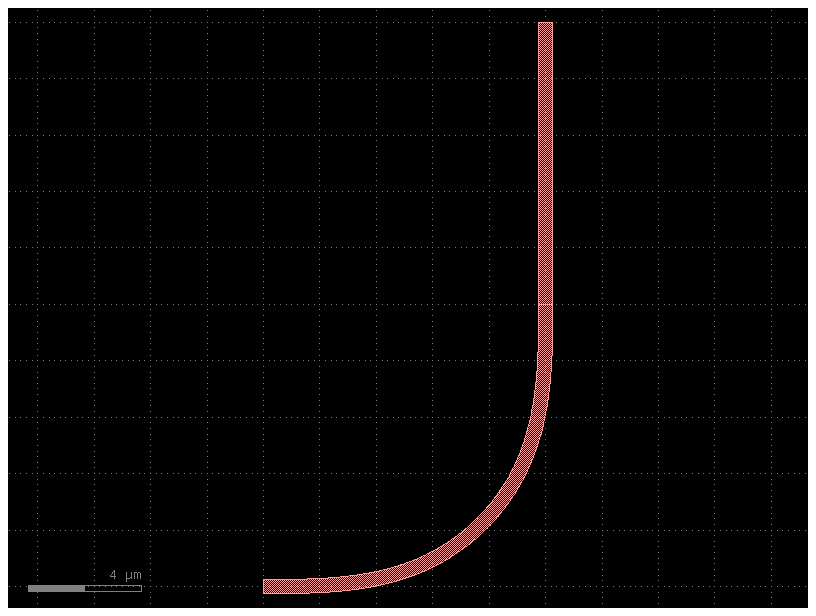
c = bend_with_straight()
c.plot()
1. String#
You can use any string registered in the PDK. Go to the PDK tutorial to learn how to register cells in a PDK.
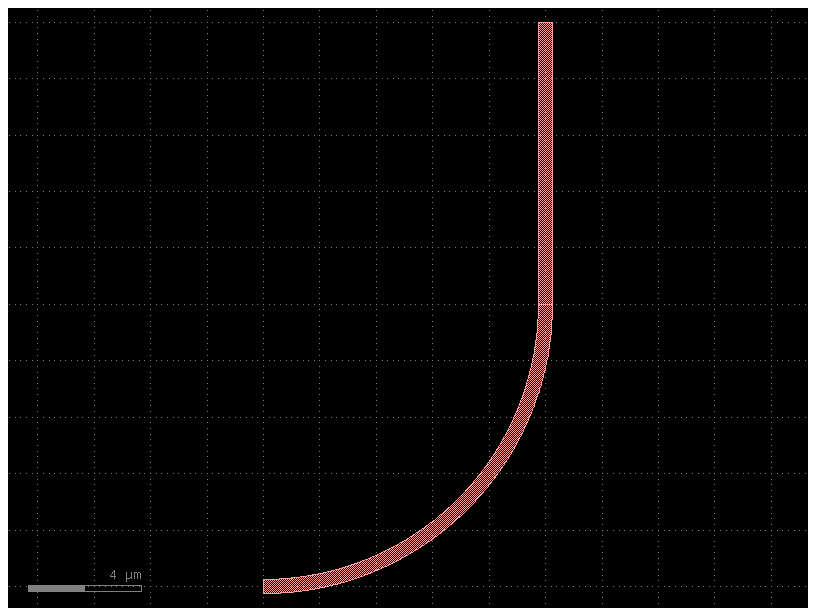
c = bend_with_straight(bend="bend_circular")
c.plot()
2. Dictionary#
You can pass a dict of settings.
bend = gf.get_component("bend_circular", radius=20)
c = bend_with_straight(bend=bend)
c.plot()
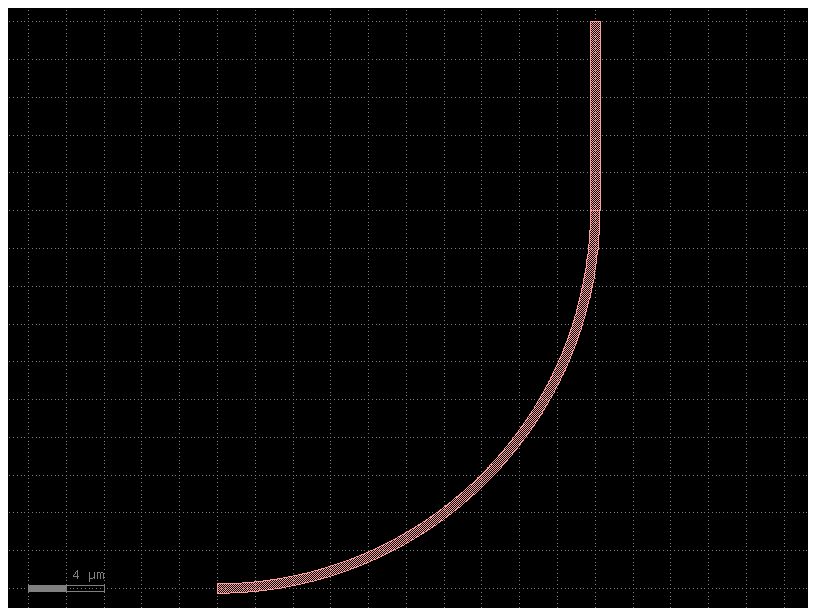
3. Function#
You can pass a function of a function with customized default input parameters from functools import partial.
Partial lets you define different default parameters for a function, so you can modify the default settings for each child cell.
c = bend_with_straight(bend=partial(gf.components.bend_circular, radius=30))
c.plot()
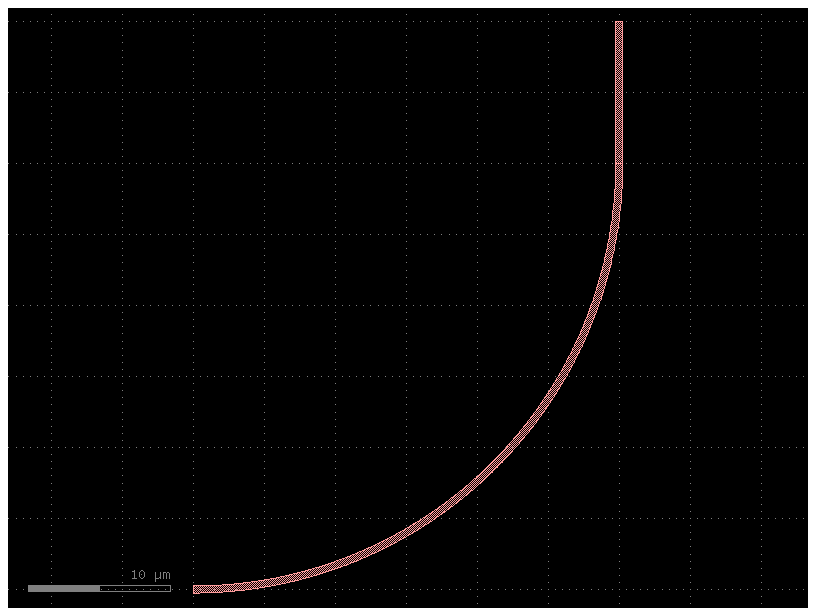
bend20 = partial(gf.components.bend_circular, radius=20)
b = bend20()
b.plot()
type(bend20)
functools.partial
bend20.func.__name__
'bend_circular'
bend20.keywords
{'radius': 20}
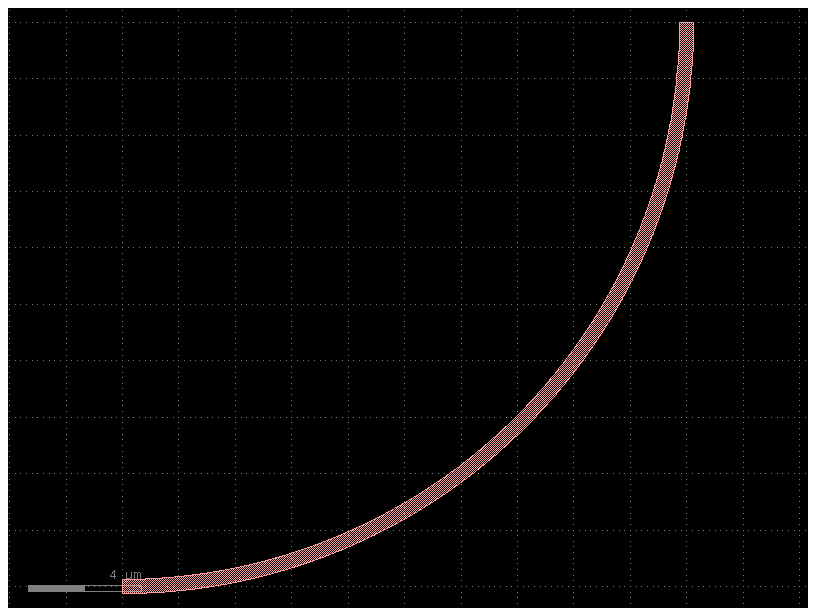
b = bend_with_straight(bend=bend20)
print(b.info.length)
b.plot()
41.416

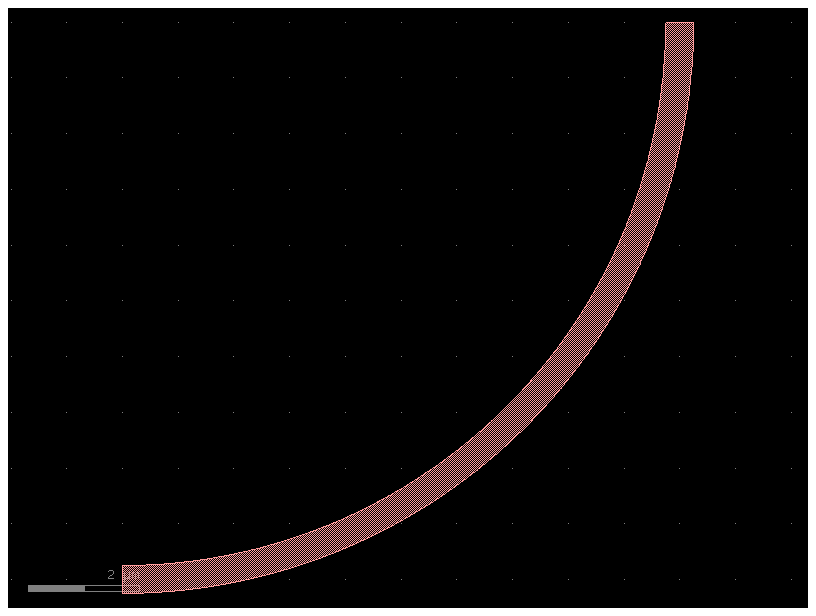
# You can still modify the bend to have any bend radius.
b3 = bend20(radius=10)
b3.plot()
Composing functions#
You can combine more complex functions out of smaller functions.
Let us say that we want to add tapers and grating couplers to a wide waveguide:
c1 = gf.components.straight()
c1.plot()

straight_wide = partial(gf.components.straight, width=3)
c3 = straight_wide()
c3.plot()

c1 = gf.components.straight()
c1.plot()

c2 = gf.c.extend_ports(c1, length=5)
c2

c3 = gf.routing.add_fiber_array(c2, with_loopback=False)
c3.plot()
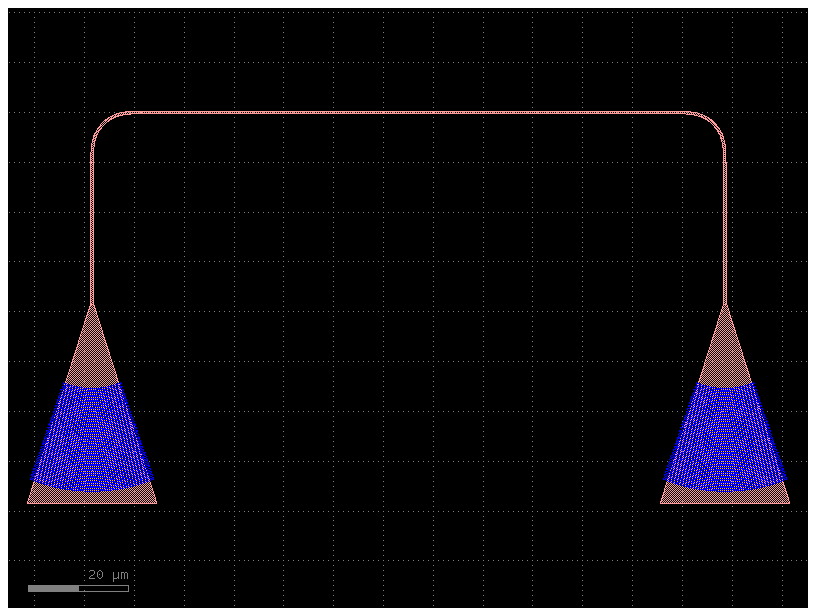
Now we do it with a single step thanks to toolz.pipe.
# The partial function is used to create simplified, pre-configured versions of other functions.
# add_fiber_array: A new version of the function that adds grating couplers, but with the with_loopback option always set to False.
# add_tapers: A new function that extends ports, but is pre-configured to always use the defined taper component as the extension.
add_fiber_array = partial(gf.routing.add_fiber_array, with_loopback=False)
c1 = gf.c.straight(width=5)
taper = gf.components.taper(length=10, width1=5, width2=0.5)
add_tapers = partial(gf.c.extend_ports, extension=taper)
# Pipe is more readable than the equivalent add_fiber_array(add_tapers(c1)).
# The toolz.pipe function takes an initial object (c1) and "pipes" it through a series of functions. The output of one function becomes the input for the next.
c3 = toolz.pipe(c1, add_tapers, add_fiber_array)
c3.plot()
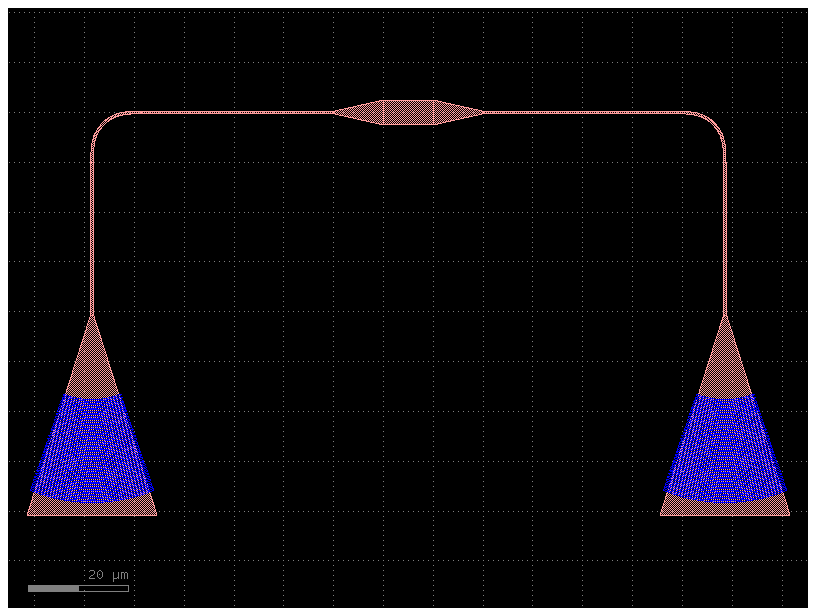
we can even combine add_tapers and add_fiber_array thanks to toolz.compose or toolz.compose
For example:
add_tapers_fiber_array = toolz.compose_left(add_tapers, add_fiber_array)
c4 = add_tapers_fiber_array(c1)
c4.plot()

is equivalent to:
c5 = add_fiber_array(add_tapers(c1))
c5.plot()

which is the same as:
add_tapers_fiber_array = toolz.compose(add_fiber_array, add_tapers)
c6 = add_tapers_fiber_array(c1)
c6.plot()

or:
c7 = toolz.pipe(c1, add_tapers, add_fiber_array)
c7.plot()

